Cas9 and DNA identification
This subpage constitutes the first part of the theory for Biotech Academy’s material on CRISPR-Cas9.
Genetic modification and DNA doublestrand breaking


This subpage constitutes the first part of the theory for Biotech Academy’s material on CRISPR-Cas9.
The Cas9 protein forms precise double-strand breaks in self-selected positions in DNA. The location of the double-strand break depends on the selected gRNA, which is bound in Cas9. Double strand breaks are obvious options for modifying the DNA sequence. It is either possible to insert sequences into double-strand breaks or make mutations in the sequence, which is carried out by exploiting the cell’s own DNA repair mechanisms. The genetic modification itself therefore lies in how to exploit the double-strand fractures. The exact location of the double strand breaks is essential for the result, as you want to target specific genes or regulatory sequences in the DNA.
Double strand breaks in DNA are a very serious situation for a cell if they are not created during the cell’s own controlled processes. A double-strand break separates the genome physically, as both DNA strands are broken, and can have serious consequences, such as cell death. Therefore, the cell will do everything possible to reassemble the DNA with its repair mechanisms. The two main types of repair systems are non-homologous end joining (NHEJ) and homology directed repair (HDR), each of which repairs double-strand breakage by its own mechanism.
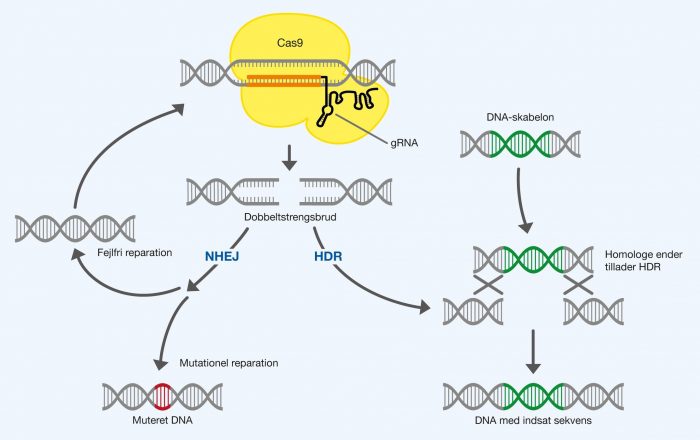
Figure 1. Cas9 forms double-strand breaks in DNA from the location chosen with the bound gRNA. Double strand breakage is repaired either by non-homologous end joining (NHEJ) or homology directed repair (HDR). These repair DNA by two different mechanisms and therefore different results can be achieved. NHEJ collects the ends of DNA, but can make mistakes that form mutations. If NHEJ is done correctly, Cas9 will simply be able to recognize the DNA sequence again and form a new fracture. This repeats itself until a mutation is formed. HDR is different as the mechanism uses a DNA template to repair the fracture. The DNA template is usually a copy of the original sequence. However, you can determine the content of the DNA template yourself by cheating the repair system, with which you can insert self-selected DNA sequences. This requires the DNA template to have homologous ends for the double-strand break. These two repair systems are used for genetic modification in the position where you have placed your double-strand fracture.
Non-homologous end joining (NHEJ) is a repair mechanism that joins the two loose ends of the DNA strands after a double-strand break. The DNA ends are composed of the enzyme DNA ligase IV in humans. This repair mechanism is very effective, but can cause mutations. These mutations can be both insertions (insertion) and deletions (removal) of base pairs, both of which have the potential to cause frame-shift mutations. Protein-coding DNA is read as codons, which are ranges of 3 nucleotides. A change that does not follow this system will interfere with the interpretation of the code. The DNA reading frame is shifted by frame-shift mutations as base pairs are inserted or removed in a number that is not a multiple of 3. It may also happen that the frame-shift mutation happens to introduce a stop codon into the code that stops the protein translation prematurely. The mutations mean that the DNA sequence can no longer be read correctly and that the protein that the sequence encodes will become dysfunctional.
The repair is likely to be successful in most cases. The important point here is that Cas9 can recognize the repaired sequence again and make a new double-strand break, which in turn can be repaired by NHEJ. This process will repeat itself until the NHEJ mistakenly makes a mutation in the DNA sequence. Then Cas9 will no longer be able to recognize the DNA sequence, as it is permanently altered with a mutation. The smart thing is that the mutation has been introduced at the position, which was determined by the self-selected gRNA. In this way, Cas9 can be used to form permanent mutations in self-selected positions in a DNA sequence. This is illustrated in Figure 1.
Homology directed repair (HDR) is a more accurate and complicated repair mechanism that uses a DNA sequence as a template to repair a double-strand break. Here, a DNA sequence is inserted into the fracture, which is similar to the template used. The DNA template must have ends that are homologous to the ends of the double-strand break. This means that the ends have overlapping and similar DNA sequences between the two pieces of DNA. This is smart for the natural use of the repair system, as multiple copies of the same DNA sequences are often found in the cell. In this way, a piece of identical DNA can be found from the homologous ends and serve as a template for the recreation of the original sequence after a breakup.
HDR can only base its repair on the basis of the homologous ends of the DNA template, but not what’s in between. Therefore, one can force the repair system to insert self-selected DNA sequences, as long as they have homologous ends. When you want to insert a sequence into a specific position in the DNA, you first form the double strand break with your Cas9. By then introducing an artificial DNA template with homologous ends, one can insert the sequence into the fracture using the repair mechanism. This is illustrated in Figure 1.
In short, it can be said that NHEJ can collect the DNA ends and possibly introduce small random mutations. HDR can restore the original DNA sequence or introduce a new DNA sequence depending on a DNA template with homologous ends to the double-strand break.
The CRISPR/Cas9 genetic modification system needs only two components to be active, namely the Cas9 protein and the selected gRNA.
The Cas9 protein targets specific DNA sequences using gRNA, which is bound in Cas9 and interchangeable. This gRNA has a sequence of 20 nucleotides at one end that is complementary to the targeted DNA sequence, which is similarly also 20 nucleotides long.
The 20 nucleotides in gRNA recognize the DNA sequences by binding to them and therefore gRNA determines which DNA sequence Cas9 is looking for. In this way, Cas9 can be reprogrammed to recognize different DNA sequences by selecting the sequence of the bound gRNA.
There is an additional requirement beyond the gRNA sequence. The targeted DNA sequence must contain a PAM sequence. This is 5′-NGG-3′ for Cas9 from S. pyogenes, where the first base (N) can be any base, followed by 2 guanine bases in the 5′ to 3′ direction of the DNA strand.
The PAM sequence always lies on the opposite string to the string on which the 20 nucleotides that are recognized lie. The 20 nucleotides must be a direct extension of the PAM sequence.
Figure 2 shows how to target different DNA sequences by replacing gRNA in Cas9 and positioning the PAM sequence in relation to the targeted 20 nucleotides.
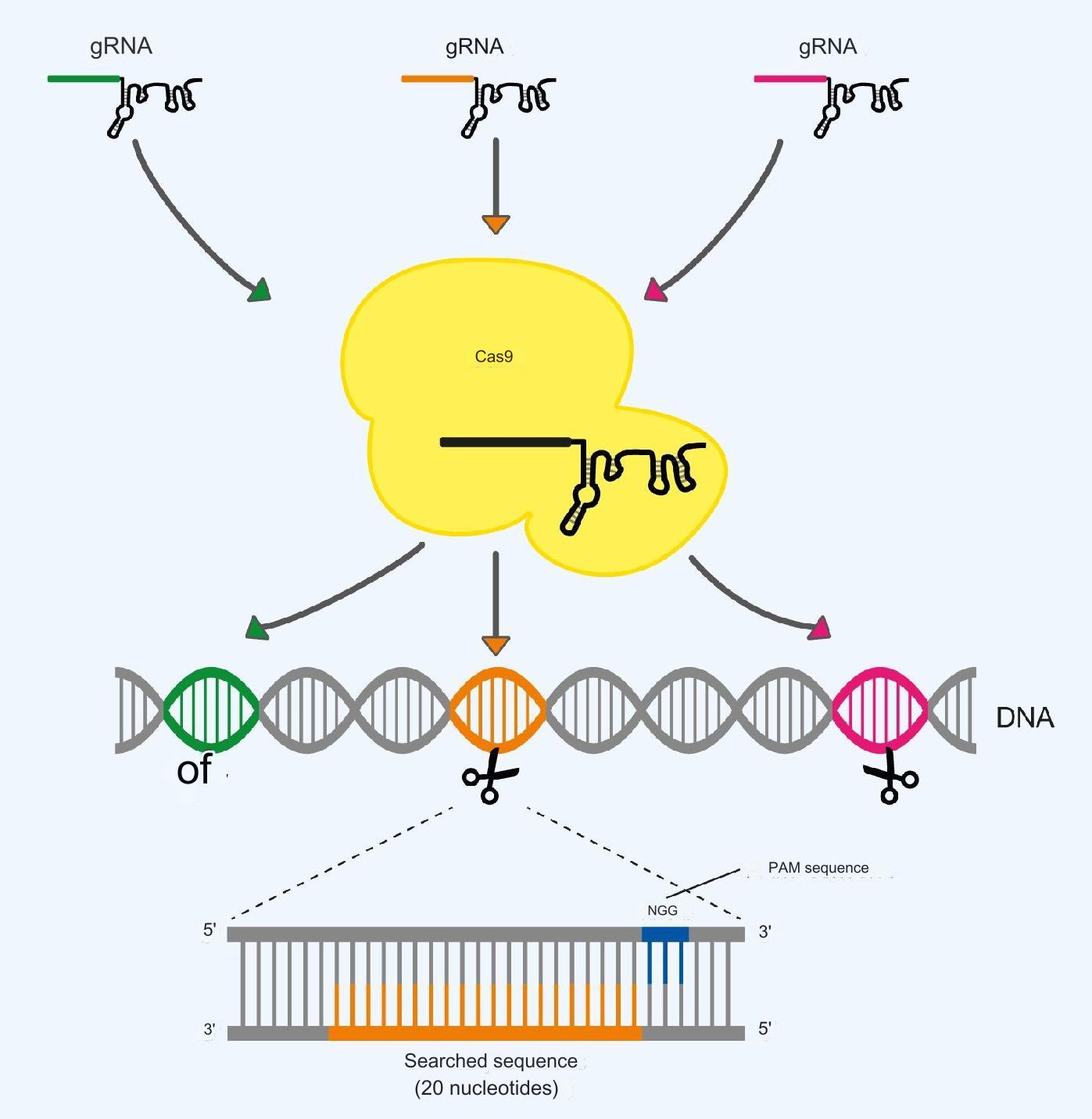
Figure 2. Reprogramming Cas9 with different gRNA. The Cas9 protein recognizes different DNA sequences according to the 20 nucleotides in the bound gRNA. A 5′-NGG-3′ PAM sequence must be present on the opposite strand next to the 20 wanted nucleotides for recognition of the DNA sequence to take place.
The protein consists of two main parts, the identification part (REC), which is responsible for the identification of the specific DNA sequence, and the nuclease part (NUC), which is responsible for the cleavage of the DNA sequence. A PAM interacting (PI) domain sits in NUC and is part of Cas9 that recognizes the PAM sequence 5′-NGG-3′. REC maintains gRNA at one end, so that the recognizing 20 nucleotides at the other end are exposed on the surface of Cas9, allowing the identification of the specific DNA sequence. Once gRNA has bound to the DNA sought, it is at the interface between REC and NUC, as can be seen in Figure 3.
The fact that Cas9 is an endonuclease means that it can split in the middle of DNA and not just at the ends. In NUC, there are two endonuclease domains, each cleaving their own strand of the wanted DNA. The HNH domain cleaves the DNA strand that contains the sequence sought by gRNA. The RuvC domain cleaves the opposite DNA strand, which contains the PAM sequence. Together, the two endonucleases form the precise double-strand break in the DNA.
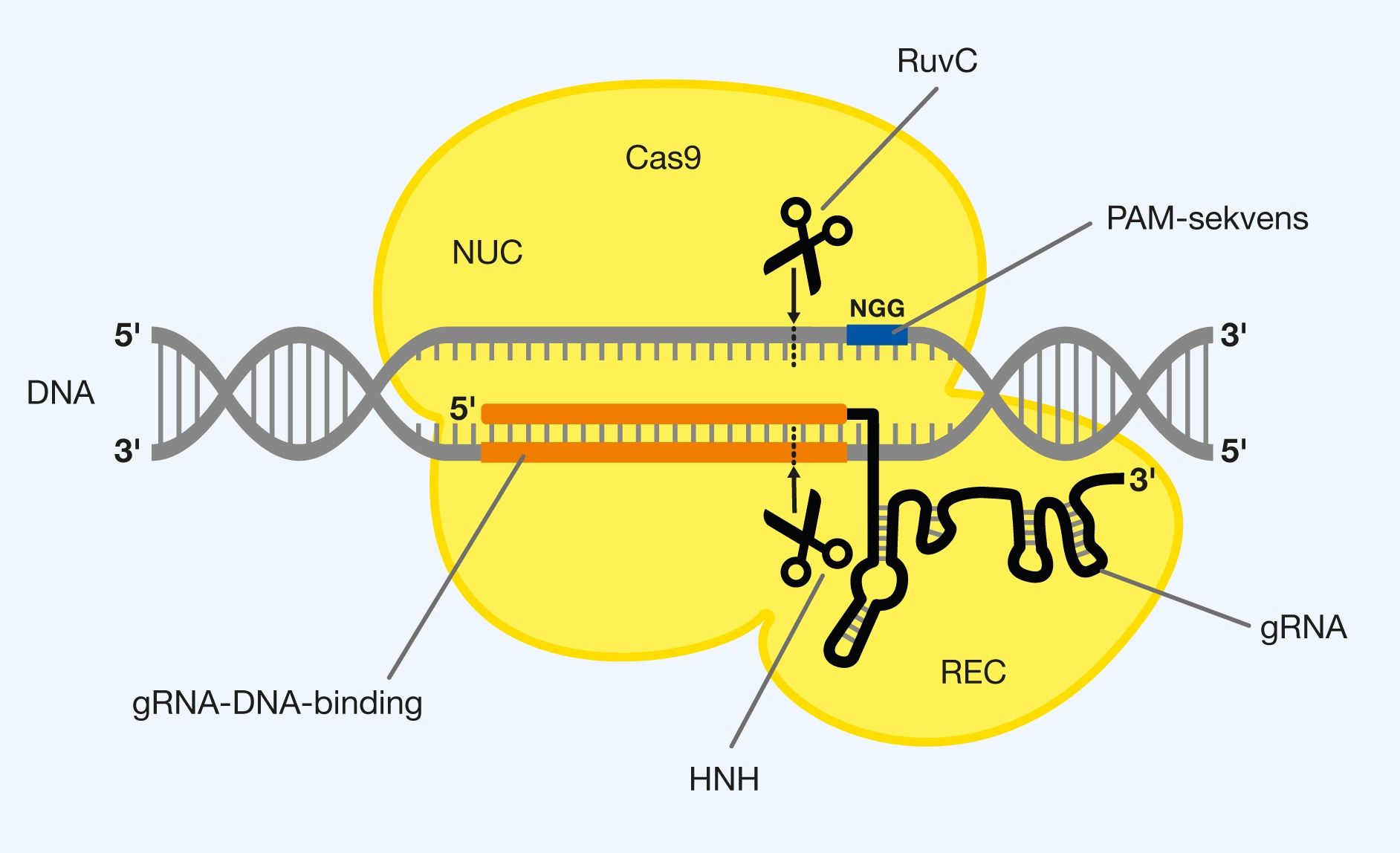
Figure 3. The structure of Cas9 with gRNA. Cas9 consists of 2 main parts, called REC and NUC. gRNA is bound in the REC, which stands for identifying DNA sequences. NUC contains a domain that recognizes the PAM sequence (5′-NGG-3′). NUC stands for the cleavage of DNA and has 2 endonuclease domains, of which RuvC cuts the DNA strand with the PAM sequence and HNH cuts the DNA strand recognized by gRNA. The double-strand break is formed between 3. and 4. bp after the PAM sequence, inside the recognized sequence.
This complex protein structure thus forms a biochemical apparatus designed to accurately identify DNA sequences and then cleave them precisely.
Now that the Cas9 protein structure has been described, a closer look can be taken at how the individual parts are utilized in the formation of a double-strand break in a specific DNA sequence. This interaction is very important for Cas9, as it is the one that is exploited when using Cas9 as a genetic engineering tool. The following steps describe the identification and cleavage of DNA, as well as the requirements that must be met for this to be possible.
Summary of Cas9 interaction with DNA
The most important information for the formation of double-strand fractures with Cas9 can be seen in Figure 4.
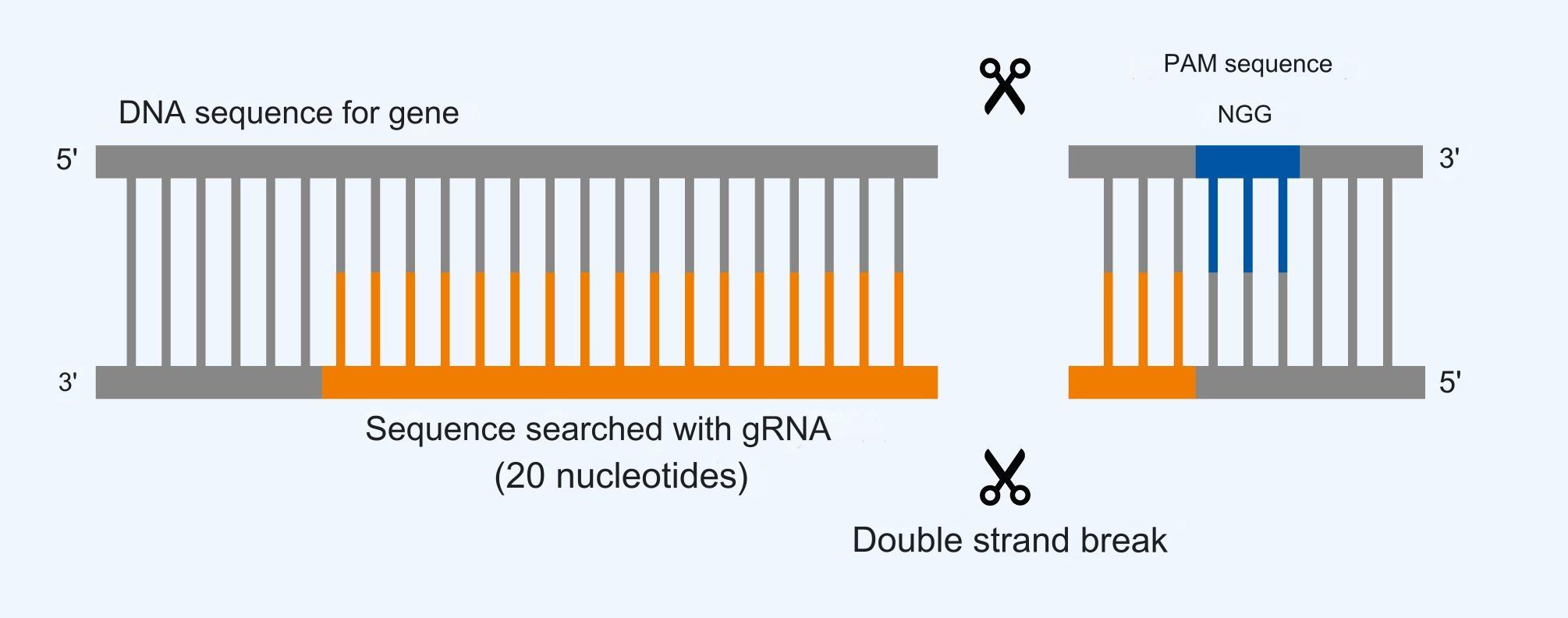
Figure 4. The important elements for Cas9 in the recognition and cleavage of a specific DNA sequence. The presence of the PAM sequence 5′-NGG-3′ allows the subsequent 20 nucleotides on the opposite strand to be recognized by the bound gRNA in Cas9. Cas9 forms the double-strand break between the 3. and 4th base pair after the PAM sequence inside the recognized sequence.
Cas9 can be used in many different ways, resulting in different molecular changes to the DNA sequence. A closer look will now be taken at some of the basic strategies that the tool can be used for and what the resulting DNA sequences will look like. The different strategies depend on how you have designed your gRNA and what repair mechanism you use. By varying which components are used, different results of the genetic modification can be achieved. For example, it is possible to utilize several pieces of gRNA. This tactic is called multiplexing and allows you to modify multiple DNA sequences at once or cut out large pieces of DNA. If you add a DNA template, you can achieve insertions of DNA sequences.
The molecular strategies are defined by how the DNA sequence is affected.
Destruction
Destruction of the gene structure by small insertions or deletions (collectively called indels) in the DNA sequence. Cas9 is used with gRNA to select the location of the double-strand fracture, after which an indel occurs as a result of mutational NHEJ repair of the fracture. Multiplexing with several pieces of gRNA can be used to destroy the same gene or different genes at once. See Figure 5.
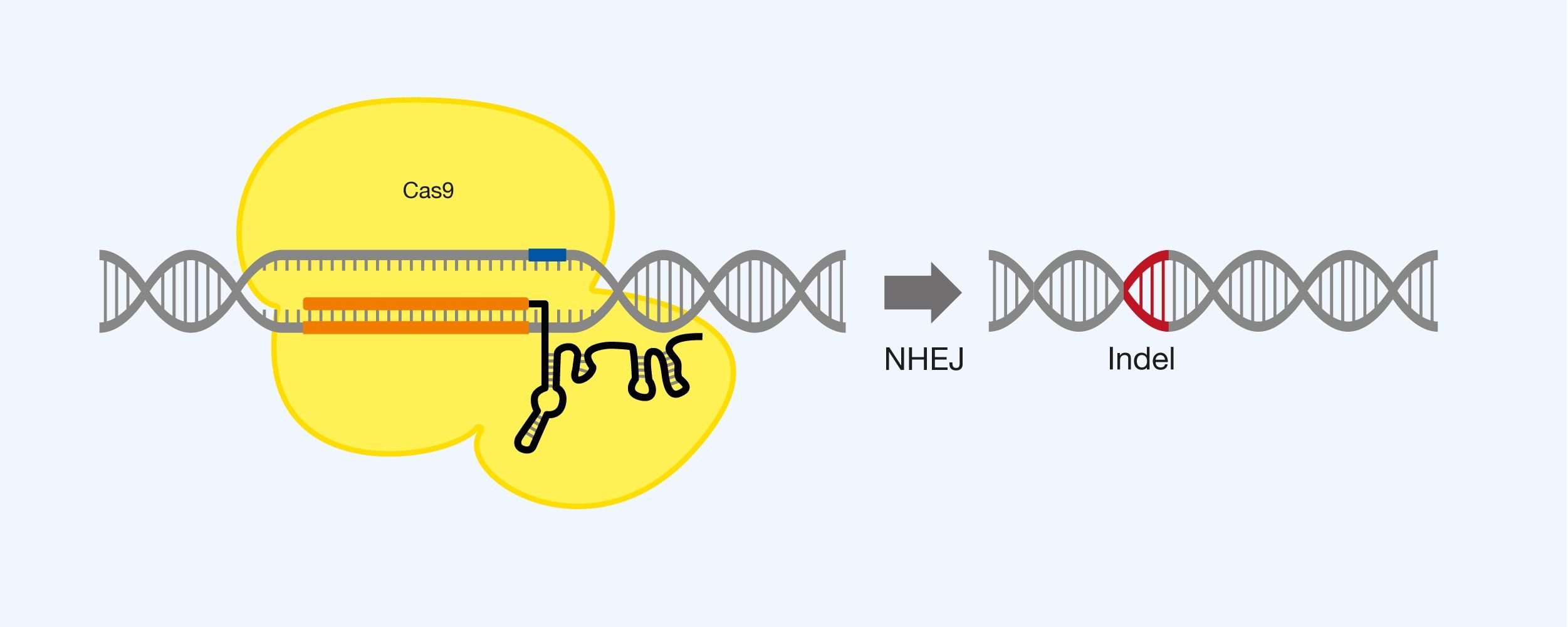
Figure 5. Destruction of a DNA sequence using Cas9 and gRNA, as well as utilization of non-homologous end joining (NHEJ).
Insertion
Insertion of larger sequences into the genome. Cas9 is used in conjunction with one gRNA to find the insertion point, and a DNA template is also introduced for insertion using HDR. If the sequence is inserted in the middle of another sequence, that sequence is likely to be destroyed. See Figure 6.
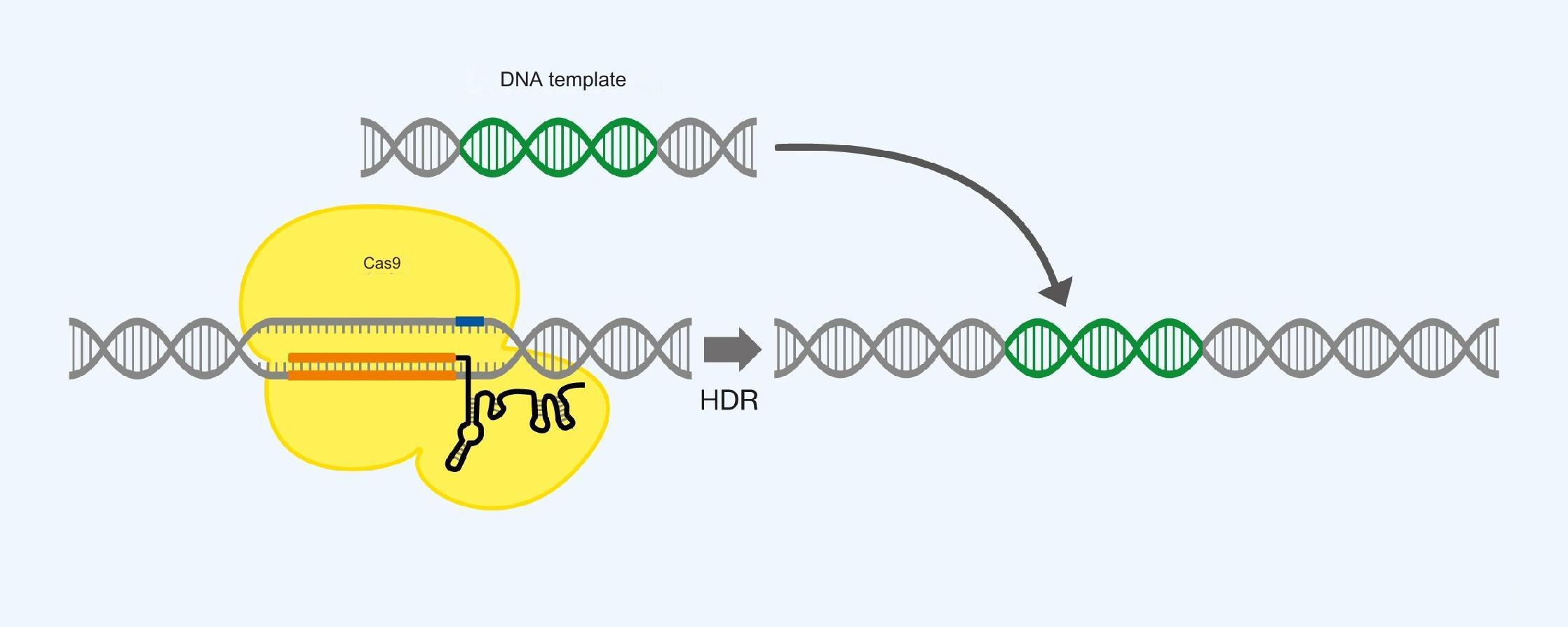
Figure 6. Insertion of a DNA sequence using Cas9, gRNA and a DNA template, as well as utilization of homology directed repair (HDR).
Excision
Deletion of larger sequences. Cas9 is used together with two pieces of gRNA, each marking the endpoints of the sequence you want to remove. For example, after the excision of the sequence, the larger fracture can be repaired by the NHEJ, which brings together the two endpoints. See Figure 7.
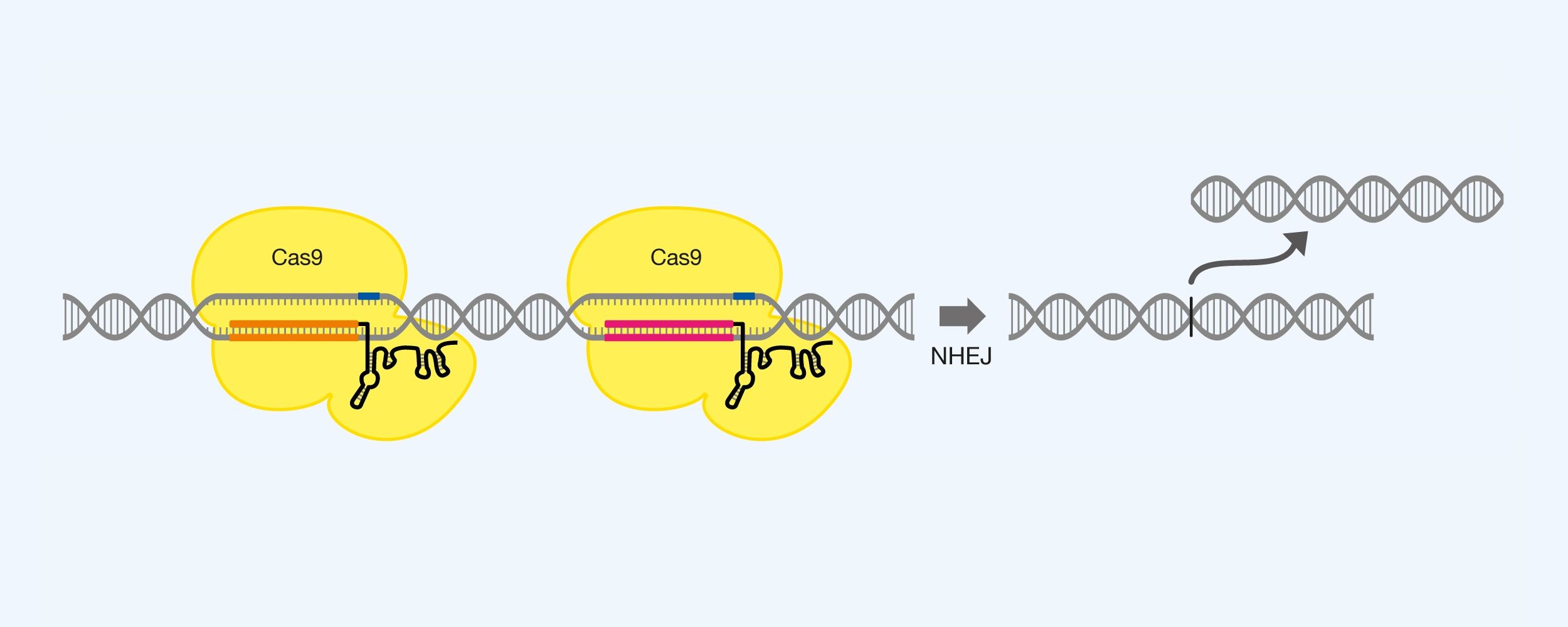
Figure 7. Excision of a DNA sequence using Cas9 and two pieces of gRNA, as well as exploitation of non-homologous end joining (NHEJ).
Substitute
Larger sequence is substituted for another sequence. Cas9 is used with two gRNAs, each marking the endpoints of the sequence to be replaced, as done by excision. A DNA template is introduced, just like insertion, which is inserted into the larger break by exploiting HDR. See Figure 8.
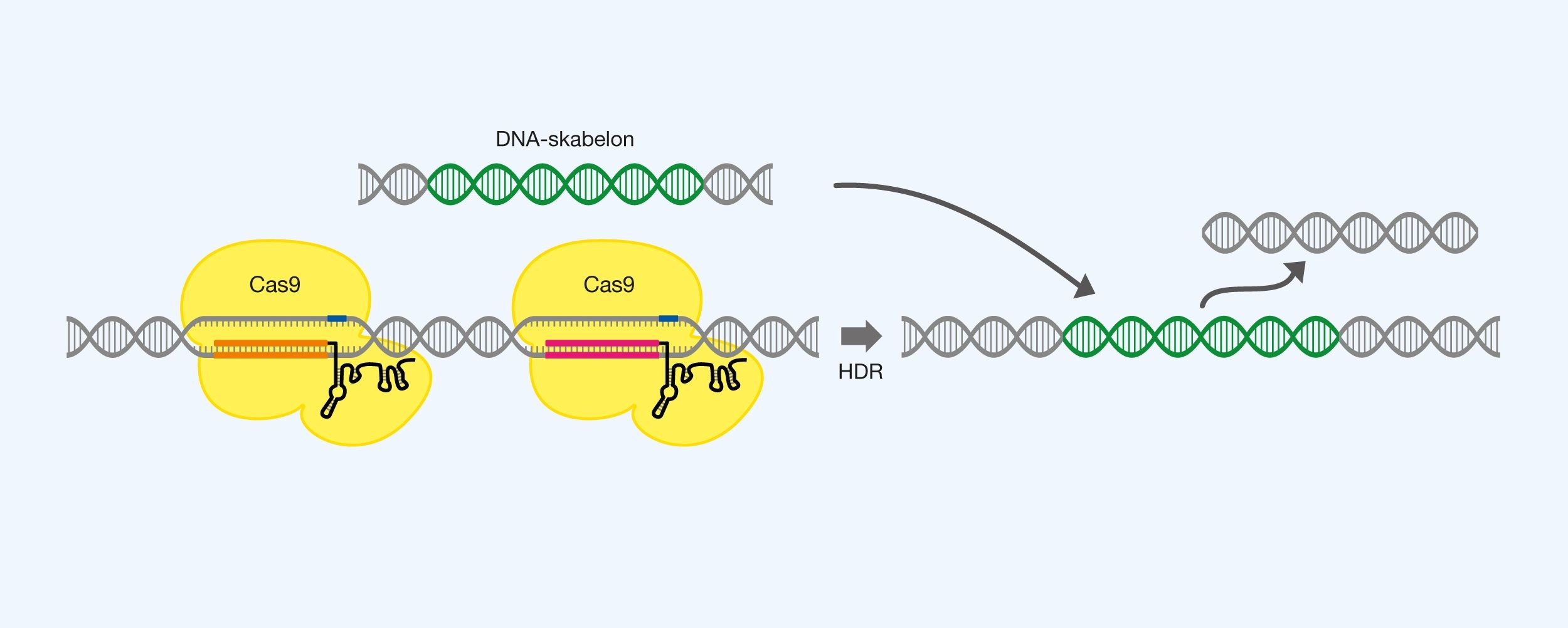
Figure 8. Replacing a DNA sequence using Cas9, two pieces of gRNA and a DNA template, as well as leveraging homology directed repair (HDR).